Experiment 2.10: Delete only red without "desaturated" label
In Ex 2.9 we succeed in deleting red without deleting cyan or other colors, with precision similar to an intervention with a cosine falloff. In that experiment, we used a subspace regularizer to attract desaturated colors to the last three dimensions. Let's see if we can get similar results without that label. We'll also try removing the unitarity regularizer to verify that it's still needed.
Hypothesis
If we weakly repel all embeddings from the anchor dimension, then we will be able to delete red without also deleting other colors. We should see error vs. color curves similar to those achieved in 2.9.
from __future__ import annotations
nbid = '2.10' # ID for tagging assets
nbname = 'Ablate red (only), 5D, fewer labels'
experiment_name = f'Ex {nbid}: {nbname}'
project = 'ex-preppy'
# Basic setup: Logging, Experiment (Modal)
import logging
import modal
from infra.requirements import freeze, project_packages
from mini.experiment import Experiment
from utils.logging import SimpleLoggingConfig
logging_config = (
SimpleLoggingConfig()
.info('notebook', 'utils', 'mini', 'ex_color')
.error('matplotlib.axes') # Silence warnings about set_aspect
)
logging_config.apply()
# This is the logger for this notebook
log = logging.getLogger(f'notebook.{nbid}')
run = Experiment(experiment_name, project=project)
run.image = modal.Image.debian_slim().pip_install(*freeze(all=True)).add_local_python_source(*project_packages())
run.before_each(logging_config.apply)
None # prevent auto-display of this cell
Model parameters
Like Ex 2.9, we use the following regularizers:
- Anchor: pins
redto $(1,0,0,0,0)$ (5D) - AxisAlignedSubspace: repels everything from dimension $1$ (with varying weight, see schedule)
- Separate: angular repulsion to reduce global clumping (applied within each batch)
Since we're isolating red, we have 5D latent embeddings and two nonlinear activation functions in the encoder and decoder, to allow the latent space to be warped more.
But unlike 2.9:
- Anti-anchor: has been removed, relying on anti-subspace to keep other concepts clear of the dimension to be ablated.
- Unitarity: is present in this list, but we'll do a run without it too.
import torch
from ex_color.loss import AngularAnchor, AxisAlignedSubspace, Separate, RegularizerConfig
from ex_color.training import TrainingModule
K = 5 # bottleneck dimensionality
N = 2 # number of nonlinear layers
RED = (1, 0, 0, 0, 0)
reg_separate = RegularizerConfig(
name='reg-separate',
compute_loss_term=Separate(power=100.0, shift=True),
label_affinities=None,
layer_affinities=['bottleneck'],
)
reg_anchor = RegularizerConfig(
name='reg-anchor',
compute_loss_term=AngularAnchor(torch.tensor(RED, dtype=torch.float32)),
label_affinities={'red': 1.0},
layer_affinities=['bottleneck'],
phase=('train', 'validate'),
)
reg_anti_subspace = RegularizerConfig(
name='reg-anti-subspace',
compute_loss_term=AxisAlignedSubspace((0,), invert=True),
label_affinities=None,
layer_affinities=['bottleneck'],
)
from typing import cast
import matplotlib.pyplot as plt
from matplotlib.axes import Axes
from IPython.display import display, Markdown
from mini.temporal.dopesheet import Dopesheet
from mini.temporal.timeline import Timeline
from mini.temporal.vis import plot_timeline, realize_timeline, ParamGroup
from utils.nb import displayer_mpl
from utils.plt import Theme
dopesheet = Dopesheet.from_csv(f'./ex-{nbid}-dopesheet.csv')
display(Markdown(f"""## Parameter schedule \n{dopesheet.to_markdown()}"""))
def plot_dopesheet(dopesheet: Dopesheet, theme: Theme):
timeline = Timeline(dopesheet)
history_df = realize_timeline(timeline)
keyframes_df = dopesheet.as_df()
fig = plt.figure(figsize=(9, 3), constrained_layout=True)
axs = fig.subplots(2, 1, sharex=True, height_ratios=[3, 1])
ax1, ax2 = cast(tuple[Axes, ...], axs)
plot_timeline(
history_df,
keyframes_df,
groups=(ParamGroup(name='', params=[p for p in dopesheet.props if p not in {'lr'}]),),
theme=theme,
ax=ax1,
show_phase_labels=False,
)
ax1.set_ylabel('Weight')
ax1.set_xlabel('')
plot_timeline(
history_df,
keyframes_df,
groups=(ParamGroup(name='', params=['lr']),),
theme=theme,
ax=ax2,
show_legend=False,
show_phase_labels=False,
)
ax2.set_ylabel('LR')
# add a little space on the y-axis extents
ax1.set_ylim(ax1.get_ylim()[0] * 1.1, ax1.get_ylim()[1] * 1.1)
ax2.set_ylim(ax2.get_ylim()[0] * 1.1, ax2.get_ylim()[1] * 1.1)
return fig
with displayer_mpl(
f'large-assets/ex-{nbid}-dopesheet.png',
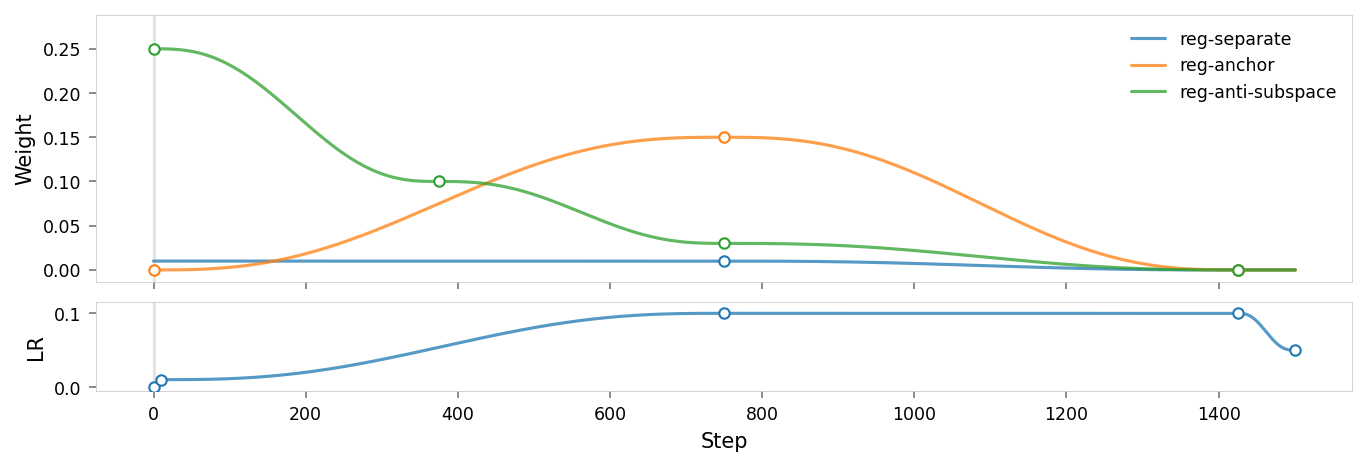
alt_text="""Plot showing the parameter schedule for the training run, titled "{title}". The plot has two sections: the upper section shows various regularization weights over time, and the lower section shows the learning rate over time. The x-axis represents training steps.""",
) as show:
show(lambda theme: plot_dopesheet(dopesheet, theme))
from torch import Tensor
from torch.utils.data import DataLoader, TensorDataset, RandomSampler
import numpy as np
from ex_color.data.color_cube import ColorCube
from ex_color.data.cube_dataset import CubeDataset, redness, stochastic_labels, exact_labels
# from ex_color.data.cube_sampler import vibrancy
from ex_color.data.cyclic import arange_cyclic
def prep_data() -> DataLoader:
cube = ColorCube.from_rgb(
r=np.linspace(0, 1, 10),
g=np.linspace(0, 1, 10),
b=np.linspace(0, 1, 10),
)
# Softly label _red_ - will be stochastically discretized in the dataloader
cube = cube.assign(
red=redness(cube['color']) ** 8 * 0.08,
# vibrant=vibrancy(cube['color']) ** 100 * 0.02,
# desaturated=(1 - vibrancy(cube['color'])) ** 10 * 0.02,
)
dataset = CubeDataset(cube)
return DataLoader(
dataset,
batch_size=64,
num_workers=4,
sampler=RandomSampler(dataset, num_samples=len(dataset), replacement=True),
collate_fn=stochastic_labels,
)
def prep_val_data() -> DataLoader:
cube = ColorCube.from_rgb(
r=np.linspace(0, 1, 4),
g=np.linspace(0, 1, 4),
b=np.linspace(0, 1, 4),
)
# Exact labels for validation: we only check where the prototypes are located
cube = cube.assign(
red=redness(cube['color']) == 1,
# vibrant=vibrancy(cube['color']) == 1,
)
dataset = CubeDataset(cube)
return DataLoader(
dataset,
# batch_size=len(dataset),
num_workers=2,
collate_fn=exact_labels,
)
import numpy as np
from ex_color.data.color_cube import ColorCube
from ex_color.data.cyclic import arange_cyclic
hsv_cube = ColorCube.from_hsv(
h=arange_cyclic(step_size=1 / 24),
s=np.linspace(0, 1, 4),
v=np.linspace(0, 1, 8),
).permute('svh')
hd_hsv_cube = ColorCube.from_hsv(
h=arange_cyclic(step_size=1 / 240),
s=np.linspace(0, 1, 48),
v=np.linspace(0, 1, 48),
)
hd_hsv_cube = hd_hsv_cube[::2, ::2, ::2]
rgb_cube = ColorCube.from_rgb(
r=np.linspace(0, 1, 20),
g=np.linspace(0, 1, 20),
b=np.linspace(0, 1, 20),
)
# with displayer_mpl(
# f'large-assets/ex-{nbid}-true-colors.png',
# alt_text="""Plot showing four slices of the HSV cube, titled "{title}". Each slice has constant saturation, but varies in value (brightness) from top to bottom, and in hue from left to right. The first slice shows a grayscale gradient from black to white; the last shows the fully-saturated color spectrum.""",
# ) as show:
# show(lambda: plot_colors(hsv_cube, title='True colors'))
from tempfile import gettempdir
import wandb
from ex_color.model import CNColorMLP
# @run.thither(env={'WANDB_API_KEY': wandb.Api().api_key})
async def train(
dopesheet: Dopesheet,
regularizers: list[RegularizerConfig],
k_bottleneck: int,
n_nonlinear: int,
*,
seed: int | None = None,
) -> CNColorMLP:
"""Train the model with the given dopesheet and variant."""
import lightning as L
from lightning.pytorch.loggers import WandbLogger
from ex_color.callbacks import LabelProportionCallback
from ex_color.seed import set_deterministic_mode
from utils.progress.lightning import LightningProgress
log.info(f'Training with: {[r.name for r in regularizers]}')
if seed is not None:
set_deterministic_mode(seed)
train_loader = prep_data()
val_loader = prep_val_data()
model = CNColorMLP(k_bottleneck, n_nonlinear=n_nonlinear)
total_params = sum(p.numel() for p in model.parameters() if p.requires_grad)
log.debug(f'Model initialized with {total_params:,} trainable parameters.')
training_module = TrainingModule(model, dopesheet, torch.nn.MSELoss(), regularizers)
logger = WandbLogger(experiment_name, project=project, save_dir=gettempdir())
trainer = L.Trainer(
max_steps=len(dopesheet),
callbacks=[
LightningProgress(),
LabelProportionCallback(prefix='labels', get_active_labels=lambda: training_module.active_labels),
],
enable_checkpointing=False,
enable_model_summary=False,
# enable_progress_bar=True,
check_val_every_n_epoch=10,
logger=logger,
log_every_n_steps=min(50, len(train_loader)),
)
print(f'max_steps: {len(dopesheet)}, train_loader length: {len(train_loader)}')
# Train the model
try:
trainer.fit(training_module, train_loader, val_loader)
finally:
wandb.finish()
# This is only a small model, so it's OK to return it rather than storing and loading a checkpoint remotely
return model
We wrap the model that we trained above in an InferenceModule. We won't be using its intervention features.
import torch
import numpy as np
from ex_color.inference import InferenceModule
async def infer_with_latent_capture(
model: CNColorMLP,
test_data: Tensor,
layer_name: str = 'bottleneck',
) -> tuple[Tensor, Tensor]:
module = InferenceModule(model, [], capture_layers=[layer_name])
import lightning as L
trainer = L.Trainer(enable_checkpointing=False, enable_model_summary=False, enable_progress_bar=False, logger=False)
batches = trainer.predict(
module,
DataLoader(
TensorDataset(test_data.reshape((-1, 3))),
batch_size=64,
collate_fn=lambda batch: torch.stack([row[0] for row in batch], 0),
),
)
assert batches is not None
preds = [item for batch in batches for item in batch]
y = torch.cat(preds).reshape(test_data.shape)
# Read captured activations as a flat [N, D] tensor
latents = module.read_captured(layer_name)
return y, latents
from torch.nn import functional as F
async def test(model: CNColorMLP, test_data: ColorCube) -> ColorCube:
x = torch.tensor(test_data.rgb_grid, dtype=torch.float32)
y, h = await infer_with_latent_capture(model, x, 'bottleneck')
per_color_loss = F.mse_loss(y, x, reduction='none').mean(dim=-1)
return test_data.assign(
recon=y.numpy().reshape((*test_data.shape, -1)),
MSE=per_color_loss.numpy().reshape((*test_data.shape, -1)),
latents=h.numpy().reshape((*test_data.shape, -1)),
)
# # Generate a list of dimensions to visualize
# from itertools import combinations
# [
# (
# b,
# a,
# (a + 1) % 5 if (a + 1) % 5 not in (a, b) else (a + 2) % 5,
# )
# for a, b in combinations((0, 1, 2, 3, 4), 2)
# ]
from typing import Sequence
from ex_color.vis import plot_colors, plot_cube_series, plot_latent_grid_3d_from_cube
from utils.nb import displayer_mpl
def tags_for_file(tags: Sequence[str]) -> str:
import re
tags = [re.sub(r'[^a-zA-Z0-9]+', '-', tag.lower()) for tag in tags]
return '-'.join(tags)
def visualize_reconstructed_cube(data: ColorCube, *, tags: Sequence[str] = ()):
with displayer_mpl(
f'large-assets/ex-{nbid}-pred-colors-{tags_for_file(tags)}.png',
alt_text="""Plot showing four slices of the HSV cube, titled "{title}". Nominally, each slice has constant saturation, but varies in value (brightness) from top to bottom, and in hue from left to right. Each color value is represented as a square patch of that color. The outer portion of the patches shows the color as reconstructed by the model; the inner portion shows the true (input) color.""",
) as show:
show(
lambda: plot_colors(
data,
title=f'Predicted colors · {" · ".join(tags)}',
colors='recon',
colors_compare='color',
)
)
def visualize_reconstruction_loss(data: ColorCube, *, tags: Sequence[str] = ()):
max_loss = np.max(data['MSE'])
median_loss = np.median(data['MSE'])
with displayer_mpl(
f'large-assets/ex-{nbid}-loss-colors-{tags_for_file(tags)}.png',
alt_text=f"""Line chart showing loss per color, titled "{{title}}". Y-axis: mean square error, ranging from zero to {max_loss:.2g}. X-axis: hue.""",
) as show:
show(
lambda: plot_cube_series(
data.permute('hsv')[:, -1:, :: (data.shape[2] // -5)],
data.permute('svh')[:, -1:, :: -(data.shape[0] // -3)],
data.permute('vsh')[:, -1:, :: -(data.shape[0] // -3)],
title=f'Reconstruction error · {" · ".join(tags)}',
var='MSE',
figsize=(12, 3),
)
)
print(f'Max loss: {max_loss:.2g}')
print(f'Median MSE: {median_loss:.2g}')
def visualize_latent_space(data: ColorCube, *, tags: Sequence[str] = (), dims: Sequence[tuple[int, int, int]]):
with displayer_mpl(
f'large-assets/ex-{nbid}-latents-{tags_for_file(tags)}.png',
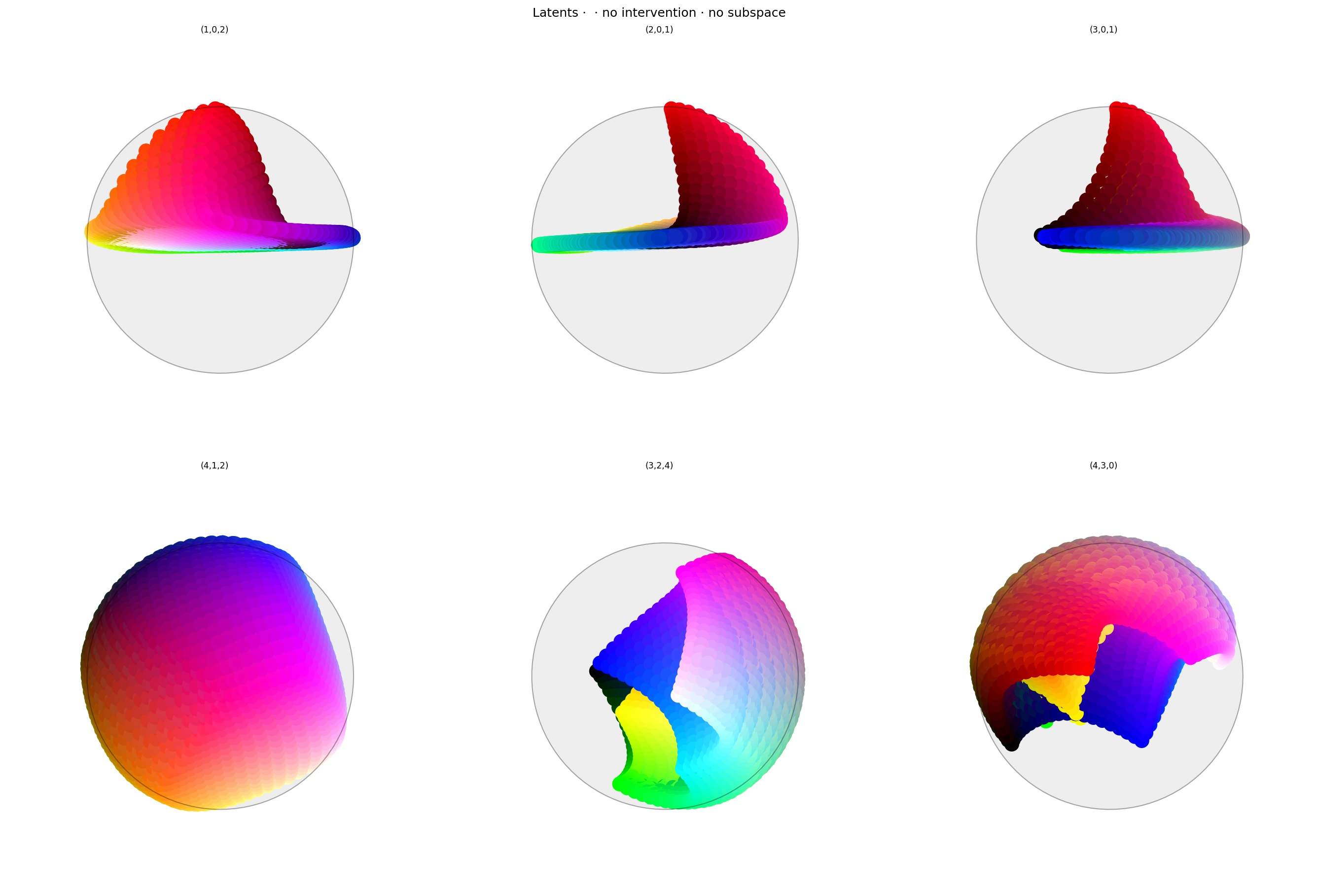
alt_text="""Two rows of three spherical plots, titled "{title}". Each plot shows a vibrant collection of colored circles or balls scattered over the surface of a hypersphere, with each plot showing one 2D projection.""",
) as show:
show(
lambda theme: plot_latent_grid_3d_from_cube(
data,
colors='recon',
colors_compare='color',
latents='latents',
title=f'Latents · · {" · ".join(tags)}',
dims=dims,
dot_radius=10,
theme=theme,
)
)
# Reload dopesheet: makes tweaking params during development easier
dopesheet = Dopesheet.from_csv(f'./ex-{nbid}-dopesheet.csv')
model = await train(
dopesheet,
[reg_separate, reg_anchor, reg_anti_subspace],
K,
N,
seed=0, # Arbitrary but not cherry-picked
)
I 6.3 no.2.10: Training with: ['reg-separate', 'reg-anchor', 'reg-anti-subspace']
INFO: Seed set to 0
I 6.3 li.fa.ut.se:Seed set to 0 I 6.3 ex.se: PyTorch set to deterministic mode
INFO: GPU available: False, used: False
I 6.4 li.py.ut.ra:GPU available: False, used: False
INFO: TPU available: False, using: 0 TPU cores
I 6.4 li.py.ut.ra:TPU available: False, using: 0 TPU cores
INFO: HPU available: False, using: 0 HPUs
I 6.4 li.py.ut.ra:HPU available: False, using: 0 HPUs max_steps: 1501, train_loader length: 16
wandb: Currently logged in as: z0r to https://api.wandb.ai. Use `wandb login --relogin` to force relogin
/tmp/wandb/run-20250921_025435-t4ogtzgh
Starting phase: Train
INFO: `Trainer.fit` stopped: `max_steps=1501` reached.
I 22.1 li.py.ut.ra:`Trainer.fit` stopped: `max_steps=1501` reached. I 22.1 ex.ca.la:Label frequencies (n=89000): _any: 0.067%, red: 0.067%
Run history:
| epoch | ▁▁▁▁▂▂▂▂▃▃▃▄▄▄▄▅▅▅▅▆▆▆▆▆▆▆▆▆▇▇▇▇▇▇▇█████ |
| labels/_any | ▁ |
| labels/epoch/_any | ▃███▇▄▅▄▅▅▃▃▄▄▃▃▃▂▂▁▁▁▁▂▂▂▁▁▁▁▂▂▂▂▂▁▁▁▁▁ |
| labels/epoch/red | ▁█▆▆▇▆▆▆▆▆▆▆▆▆▅▅▅▅▅▅▅▅▅▅▅▅▅▅▅▅▅▅▅▅▅▅▅▅▅▅ |
| labels/red | ▁ |
| train_loss | █▂▁▁▂▁▁▁▁▁▁▁▁▁▁▁▁▁▅▁▁▁▁▁▁▂▂▁▁▁▁▁▁▁▁▁▁▁▁▁ |
| train_recon | █▃▄▃▂▁▂▁▂▂▂▂▂▃▄▁▂▄▃▂▁▂▂▄▄▂▂▁▁▁▁▁▁▁▁▁▁▁▁▁ |
| train_reg-anchor | ▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁█▁▁▁▁▁▁▁▁▁▁▁▁▁ |
| train_reg-anti-subspace | ▇▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▁▂▁▂▁▁▁▁▁▁█▆▃▃▂▅▄▁▃▃▂▂▃ |
| train_reg-separate | ▅█▃▆▃▃▂▂▂▂▂▂▂▁▂▁▂▂▁▂▂▂▂▂▁▂▁▂▂▁▁▁▁▂▁▁▁▂▁▃ |
| trainer/global_step | ▁▁▁▁▂▂▂▂▂▃▃▃▃▃▃▃▃▄▄▄▄▄▄▄▅▅▅▅▅▆▆▆▆▆▇▇▇▇▇█ |
| val_loss | ▂▁▂▁█▂▁▁▁ |
| val_recon | ▂▁▂▁█▂▁▁▁ |
| val_reg-anchor | █▇█▄▄▂▁▁▁ |
Run summary:
| epoch | 92 |
| labels/_any | 0.00067 |
| labels/epoch/_any | 0.00067 |
| labels/epoch/red | 0.00067 |
| labels/red | 0.00067 |
| train_loss | 4e-05 |
| train_recon | 4e-05 |
| train_reg-anchor | 0 |
| train_reg-anti-subspace | 0.01076 |
| train_reg-separate | 0.29428 |
| trainer/global_step | 1487 |
| val_loss | 0.00013 |
| val_recon | 0.00013 |
| val_reg-anchor | 0.00021 |
View project at: https://wandb.ai/z0r/ex-preppy
Synced 5 W&B file(s), 0 media file(s), 0 artifact file(s) and 0 other file(s)
/tmp/wandb/run-20250921_025435-t4ogtzgh/logs
from IPython.display import clear_output
tags = ['no intervention', 'no subspace']
hsv_out = await test(model, hsv_cube)
hd_hsv_out = await test(model, hd_hsv_cube)
rgb_out = await test(model, rgb_cube)
clear_output()
visualize_reconstructed_cube(hsv_out, tags=tags)
visualize_reconstruction_loss(hd_hsv_out, tags=tags)
visualize_latent_space(
rgb_out,
tags=tags,
dims=[(1, 0, 2), (2, 0, 1), (3, 0, 1), (4, 1, 2), (3, 2, 4), (4, 3, 0)],
)

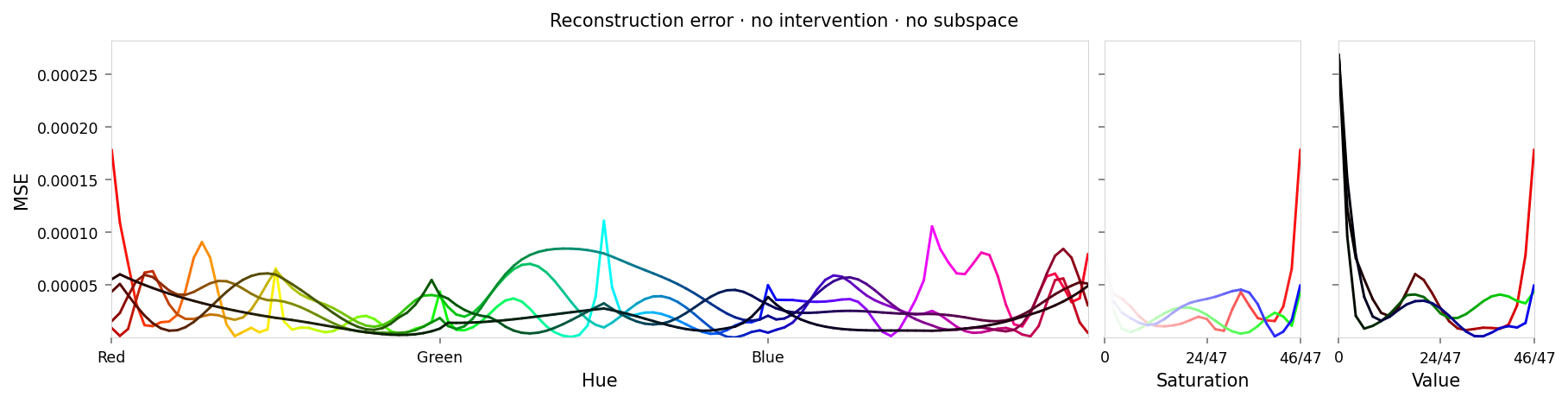
Max loss: 0.00027 Median MSE: 2.2e-05
Reconstruction loss looks about as good as last time. There's higher loss around red even without intervenion, but it's still low.
Latent space looks suprisingly good: the area opposing red is clear, and there's a pronounced collection of reds near the anchor point.
from ex_color.surgery import ablate
ablated_model = ablate(model, 'bottleneck', [0])
from IPython.display import clear_output
tags = ['ablated', 'no intervention', 'no subspace']
hsv_out = await test(ablated_model, hsv_cube)
hd_hsv_out = await test(ablated_model, hd_hsv_cube)
rgb_out = await test(ablated_model, rgb_cube)
clear_output()
visualize_reconstructed_cube(hsv_out, tags=tags)
visualize_reconstruction_loss(hd_hsv_out, tags=tags)
visualize_latent_space(
rgb_out,
tags=tags,
dims=[(1, 0, 2), (2, 0, 1), (3, 0, 1), (4, 1, 2), (3, 2, 4), (4, 3, 0)],
)

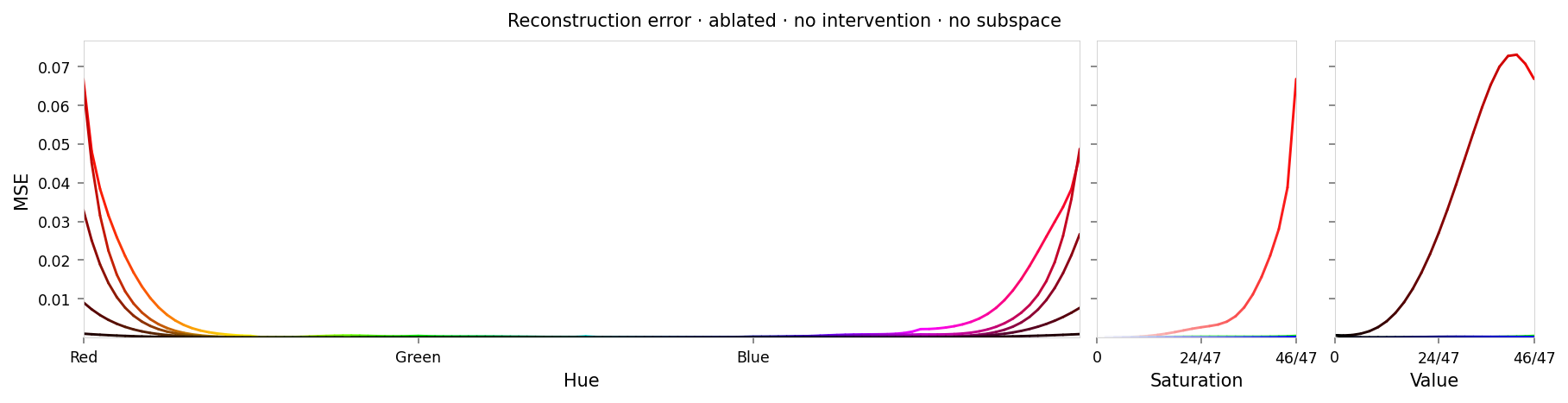
Max loss: 0.073 Median MSE: 6e-05
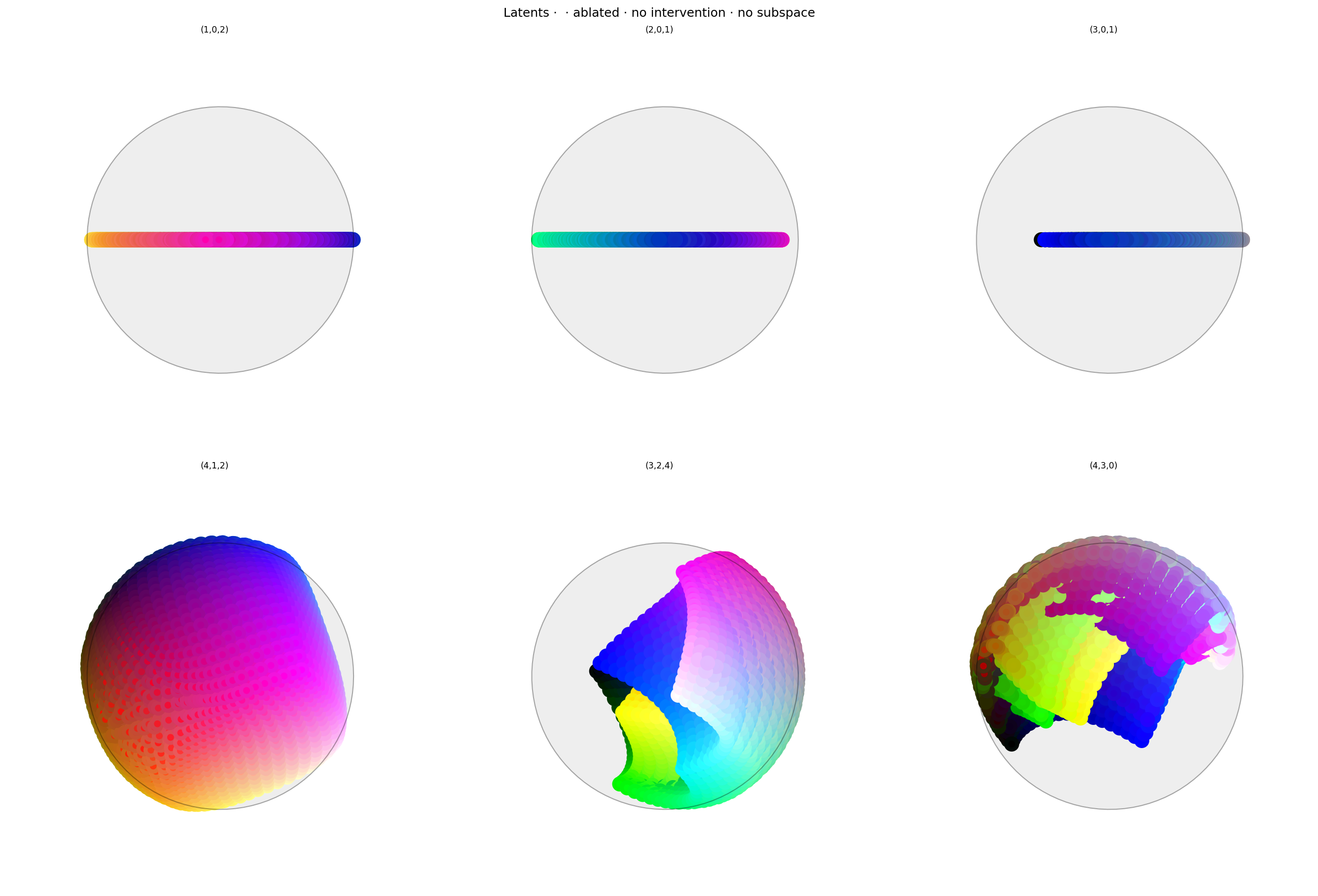
This is almost as good as last time: about 1/3 the loss for red (higher would be better), with fairly smooth falloff to the yellow and purple. There's hardly any error for other colors.
Conclusion
We can delete red without deleting cyan or other colors, even without labelling anything other than red. The error at red could be higher; perhaps that could be achieved with more hyperparameter tuning.
When we first tried this (see git history), it didn't work well: red was isolated, but the falloff toward other colors was very sharp, so there was hardly any impact on other warm colors. We fixed that by adjusting the hyperparameter schedule: instead of having Anchor and Anti-subspace in conflict the whole time, the schedule starts with a high Anti-subspace weight, and transitions to high Anchor weight around half-way through. This causes the network to section off the anchor (first) dimension at the start of training, and pulls red into that space once the manifold has already been established.